Rapid ID of Functional inhibitors of Human POLRMT by Deep Convolutional Neural Networks
At the American Chemical Society Fall 2020 Virtual Meeting & Expo, several Atomwise members and partners were selected to present their research and work. Learn what our Atoms have been working on below and visit Atomwise at ACS Fall 2020 Virtual Meeting & Expo for other presentation sessions.
 Jeff Warrington, PhD
Jeff Warrington, PhD
Atomwise
Co-Author: Noor Khalid, PhD, Rebecca Laposa, PhD
Dept. of Pharmacology and Toxicology, University of Toronto, Canada
Title: Identification of Mitochondrial RNA Polymerase Inhibitors via Deep Convolutional Neural Networks: A Therapeutic Strategy for Cancer Metabolism
Division: MEDI
Abstract
Dysregulated metabolism is a hallmark of cancer and a substantial subset of many cancers rely on oxidative phosphorylation (OXPHOS) for growth and viability, including AML, breast cancer, lung cancer, glioblastoma, ovarian and pancreatic cancer. The electron transport chain (ETC) in mitochondria contains both nuclear DNA- and mitochondrial DNA-encoded subunits. Mitochondrial DNA-encoded subunits are essential for ETC function and are transcribed by the mitochondrial RNA polymerase POLRMT. The Laposa lab has previously validated POLRMT as an anticancer target in acute myeloid leukemia models. The challenge of inhibiting POLRMT with small molecules will arise from both a) selectivity to off targets and b) transport of the drug to the mitochondria. There exist several small nucleoside / nucleotide analogues that are known to inhibit RNA polymerase. However, this target space is rife with the opportunity for off-target activities and toxicity. Therefore, we targeted a novel pocket with the aim of finding novel chemical matter. A virtual library of 7.2 million commercially available compounds was screened against POLRMT using AtomNet® model - an artificial intelligence platform that applies deep convolutional neural networks to drug discovery. The top 72 hit compounds were acquired and tested in OCI-AML2 human acute myeloid leukemia cells for a potential decrease of mitochondrial RNA levels (MT-ND1) using reverse transcriptase polymerase chain reaction (RT-PCR). OCI-AML2 cells were treated with compounds at 0.1, 1 or 10 micromolar for 72 hours, 3 days or 6 days or with the positive control POLRMT ribonucleoside analogue inhibitor, 4'-azidocytidine. Gene expression was normalized to 18S. Twelve compounds demonstrated a time-dependent decrease in mitochondrial gene expression. In summary, these data indicate that functional inhibitors of human POLRMT can be identified rapidly via deep convolutional neural networks.
Poster Presentation
On-demand Poster (click for hi-resolution)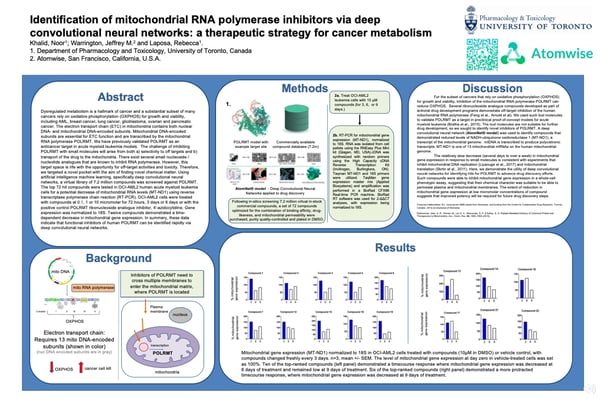
On-demand audio recording
Join our team
Our team is comprised of over 40 PhD scientists who contribute to a high-performance academic-like culture that fosters robust scientific and technical excellence. We strongly believe that data wins over opinions, and aim for as little dogma as possible in our decision making. Learn more about our team and opportunities at Atomwise.
Related Posts
Subscribe
Stay up to date on new blog posts.
Atomwise needs the contact information you provide to send you updates. You may unsubscribe from these communications at any time. For information please review our Privacy Policy.
